Instructions for Use of the MacroModel Program.
Before you start, read the general
documentation on the use of workstations. You will be using
the Silicon graphics O2 workstation in the Perkin lab and in
the main computer room, and you will need a valid user
identification and password, which should have been issued to
you when you first entered the department. After you log in,
initiate MacroModel by moving the mouse cursor over from
Applications to MacroModel and release.
After a few seconds, a mobile outline of a new window appears
on the screen which has to be "placed" somewhere on the screen
by moving the mouse cursor and clicking with the left hand
mouse button. You will probably want to place it near the top
left of the screen. Identify the terminal as 2 (3D
Silicon graphics) and press the  key. A
number of further "windows" will appear in the screen. The
largest is the molecule drawing area, and to the right of it
are the main command windows. In addition, a number of menus
"pop-up" in response to certain commands. The mouse buttons
invoke different actions on these various windows.
key. A
number of further "windows" will appear in the screen. The
largest is the molecule drawing area, and to the right of it
are the main command windows. In addition, a number of menus
"pop-up" in response to certain commands. The mouse buttons
invoke different actions on these various windows.
- The left mouse button is used to select menu items, and
also to "focus" a window, ie establish communication between
it and the keyboard from which file names etc can be
typed.
- The middle button is used to rotate the molecule in the
drawing window. The sense of rotation is selected from the
main button windows by clicking on ROT X (or Y or Z). By
default, both ROT X and ROT Y are selected, and moving the
mouse whilst holding down the middle button will rotate the
molecule about these two axes. To reset the default, "double
click" on ROT X.
- The right hand button is used to bring help. For example,
clicking with this button on a menu item opens up the Help
window, with suitable comments.
If more text than fits the size of this window is present, use
the "scroll" bars and the left hand button to view the rest of
the text;

One window always present is the message window. Here are
reported numerical results of various calculations and other
information of relevance.

The most important menus are the three shown below. You switch
from one to the other by clicking with the left button on
INPUT, ENRGY, or ANLYZ, and you will need to use them in that
order. You will only need to use a few of the menu item, but
feel free to experiment with the others!
An options menu is invoked by clicking on OPT in any of the
three main menus.You can switch ie CPK models on at any time,
but be aware it slows things down considerably;

- To start a carbon chain, click DRAW in INPUT and click on
the molecule window with the left mouse button to start a
chain. You can rotate the structure whilst building by
pressing the middle mouse button and "dragging" the mouse
around. In this way, you can impart a realistic 3D component
to the molecule you are building. Alternatively, pressing the
right mouse button enables you to "translate" the molecule
without rotating it.
- There are some organic "templates" defined on the INPUT
menu. If these are more appropriate, select one of these
instead of DRAW and position it in the middle of the screen.
Other templates can be introduced by selecting PEPTID, NUCLEI
or CARBO from the menu.
-
To continue building up a chain, move the cursor and press
the left button again. A line will appear on the screen,
representing a C-C bond. If you have a template to which
you want to attach a chain, select DRAW, and position the
cursor at the atom you want the chain added to. This may
create a pentavalent carbon, and you will have to use H DEL
(see below) to delete eg a hydrogen and H ADD to add
them. If you click once on these two menu items, the action
performed will be on a single atom only, but if you
double-click, a new menu pops up with several
options;
Quite a number of menu items are of this kind, but you only
find out which by trying.
- Continue building up the chain. If you want to close to a
cyclic compound, position the cursor accurately on the first
atom. If you want a double bond, back-track to the previously
positioned atom in the chain.
- One frequently needs to break the chain, ie to
lift the imaginary pen you are drawing with off the paper, ie
when returning to a previously defined single bond to turn it
into a double or triple bond. To do this, simply select DRAW
again. To enter carbon side-chains, first select DRAW, then
point to the carbon carrying the side chain and then start
building up a new chain. After this chain is complete, the
"pen" must be lifted off the paper as described above.
Complete all the basic skeleton of the molecule in this
manner, representing all heteroatoms by carbon at this
stage.
- To substitute any carbon atom by a heteroatom, left click
on the heteroatom in the INPUT menu, then point to the carbon
to be substituted. If you want to insert say four oxygen
atoms, point to O on the menu, then to each of the four
carbon atoms in turn.
-
To delete a specified atom, click on DELET once, then point
to the atom to be deleted, or to the middle of a bond if
you want to change a double into a single bond. To remove a
single bond, you have to first turn it into a "zero order"
bond (used for weak complexes, transition states etc) and
then delete the zero order bond. This is another
"double-clickable" item;
- If the structure you are drawing is too big for the
screen, it can be rescaled by selecting the SCALE option.
Clicking SCALE twice rescales to exactly the size of the
screen. Alternatively, if you want to magnify part of
structure, select CLIP and point the cross-hair at the bottom
left and top right of the region of the molecule to be
magnified.
- Carbon chains so drawn have no hydrogens, whereas
templates do. Very probably at this stage, the carbon
valencies are not correct, most missing hydrogen and the
templates may have too many. To ensure correct valency,
double click on H DEL and then on H ADD to ensure the correct
valencies. If these are wrong, the program will not minimise
the structure correctly.
- When the basic elements of the structure are complete,
select the ENRGY menu. Several default options are selected
automatically, including the MM2 force field. Click on
another force field if you want tho change this option.
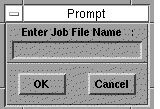
Select START to commence geometry optimisation. You will be
asked to enter a job file name. Select New and type
the name of a file, e.g. temp. Notice that the
keyboard is only active for this purpose if the cursor is
positioned over the rectangular box in which the file name is
entered and a
 type cursor
appears.
type cursor
appears.

From now on, information about the state of the optimisation
appears in the message window. If that window is obscured
(e.g. by the Options window) you may have to move it
by pressing the left mouse button on the top bar of the
window and "dragging" the menu to a new location..The
geometry on the screen will be updated as the optimisation
proceeds, giving some idea of how the shape of the molecule
is changing. If your 3D sketching has not given the program a
good enough indication of the three-dimensional structure of
the molecule, the optimisation may not converge to a sensible
answer. Alternatively, your starting point may converge to
the wrong conformation (such as boat rather than chair in a
cyclohexane ring).You have to select the INPUT option to make
appropriate alterations, and minimise again. Several cycles
of this may be needed before a satisfactory result is
obtained. You should particularly check that you have the
correct stereochemistry. For example, if you think you are
working with a cis-alkene, make sure the program has not
rearranged it to a trans-alkene when you were not looking!
There are also some 'local minima' in the MM2 or MM3
potential energy surface which have very high energies. If an
energy makes no 'chemical sense', you should look very
carefully at the geometry.
- When optimisation is complete, you are now ready to
calculate parameters such NMR coupling constants.
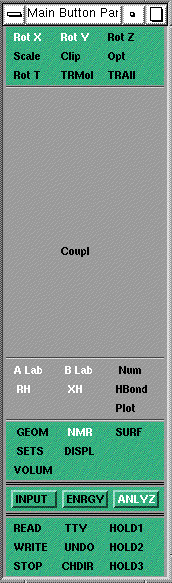
Enter the ANLYZ menu in MacroModel, then select NMR, and
finally COUPL. With the left mouse button, click at two
hydrogen atoms whose nmr coupling constant you want to
predict. The MacroModel program calculates the dihedral
angles between these hydrogens, applies the appropriate
Karplus relationship (for which a reference is given) and
displays the expected value.
Note: Record all the values in your laboratory
notebook, since there is no easy way of printing.
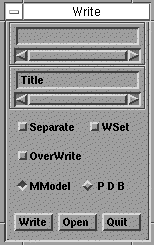
- Save your work using WRITE. Place the mouse cursor in the
first rectangular entry window (just below the Write caption)
so that a
 type cursor appears and type
a name in from the keyboard. If the mouse cursor is not
positioned there, you will not be able to enter a name. Then
click on Write.
type cursor appears and type
a name in from the keyboard. If the mouse cursor is not
positioned there, you will not be able to enter a name. Then
click on Write.
Back to Experiment



 key. A
number of further "windows" will appear in the screen. The
largest is the molecule drawing area, and to the right of it
are the main command windows. In addition, a number of menus
"pop-up" in response to certain commands. The mouse buttons
invoke different actions on these various windows.
key. A
number of further "windows" will appear in the screen. The
largest is the molecule drawing area, and to the right of it
are the main command windows. In addition, a number of menus
"pop-up" in response to certain commands. The mouse buttons
invoke different actions on these various windows.








 type cursor
appears.
type cursor
appears.



 type cursor appears and type
a name in from the keyboard. If the mouse cursor is not
positioned there, you will not be able to enter a name. Then
click on Write.
type cursor appears and type
a name in from the keyboard. If the mouse cursor is not
positioned there, you will not be able to enter a name. Then
click on Write.