Research data (and its management) is rapidly emerging as a focal point for the development of research dissemination practices. An important aspect of ensuring that such data remains fit for purpose is identifying what curation activities need to be associated with it. Here I revisit one particular case study associated with the molecular structure of a product identified from a photolysis reaction[1] and the curation of the crystallographic data associated with this study.
This particular dataset (CSD, dataDOI: 10.5517/cctnx5j) is associated with an article entitled “Single-Crystal X-ray Structure of 1,3-Dimethylcyclobutadiene by Confinement in a Crystalline Matrix“.[1] Data for crystal structures supporting a research article is required (at least in part) to be deposited into the Cambridge structure database (internal reference MUWMEX) and for which a significant level of curation is performed. Although the definition of the term curation has evolved over the last few years, here I take it to include the following:
- Identification of appropriate metadata describing the data. For molecules, this would include any identifiers such as the name of the molecule and the connectivities of the atoms constituting that molecule.
- The submission of this metadata to a suitable aggregator, such as e.g. DataCite and its inclusion in any other databases associated with the data. These two tests are part of the FAIR data guidelines[2], covering the F (findable) and A (accessible).
- Performing any validation tests for the data that can be identified. With crystal structure data in CIF format, this is defined by the utility checkCIF and helps to ensure the I (inter-operable) of FAIR. The R refers in part to the licenses under which the data can be re-used.
On (it has to be said rare) occasions, these procedures can lead to a disparity between the author’s conclusions arrived on the basis of their acquired data and the metadata identified by the independent curators. This difference is most obviously illustrated in this case study by the chemical names inferred by the curation process for the structure represented by the data in the CSD:
- chemical name: “tetrakis(Guanidinium) 25,26,27,28-tetrahydroxycalix(4)arene-5,11,17,23-tetrasulfonate 1,5-dimethyl-2-oxabicyclo[2.2.0]hex-5-en-3-one clathrate trihydrate“
- chemical name synonym: “tetrakis(Guanidinium) tetra-p-sulfocalix(4)arene 1,3-dimethylcyclobutadiene carbon dioxide clathrate trihydrate“.
Only the synonym agrees with the title given by the original authors in their publication.[1] One might indeed strongly argue that these two names are not in fact synonyms, since they refer to quite different chemical structures with different atom connectivities. A search of the database for the sub-structure corresponding to 1,3-dimethylcyclobutadiene does not reveal any hits and so the information implied by this synonym is not recorded in the index created for the CSD database.
I asked the scientific editors of the CSD for some guidance on the curation procedures applied to crystal structure datasets and they have kindly allowed me to quote some of this.
- “In cases such as this, we as editors are sometimes faced with conflicting information and have to try our best to strike a balance between the data presented in the CIF, a published interpretation and our knowledge based on the information already in the CSD”.
- “In areas where there is a particular conflict between these, we often would include a comment (usually in the Remarks or Disorder field as appropriate)”. For this particular dataset, one finds the following under the Disorder field:
- “Under UV radiation the clathrated pyrone molecule converts to a disordered mixture of square-planar 1, 3-dimethylcyclobutadiene and rectangular-bent 1, 3-dimethylcyclobutadiene in van der Waals contact with a carbon dioxide molecule. The ratio of the square-planar to rectangular-bent 1, 3-dimethylcyclobutadiene clathrate is modelled with occupancies 0.6292:0.3708”.
- It is not entirely obvious however whether this last comment originates from the original authors or from the data curators. It does not resolve the difference between the assigned chemical name and the indicated chemical name synonym.
- “In the case of MUWMEX, I think that the editor produced a diagram (below) which seems chemically reasonable based on the crystallographic data with which we were provided and tried to cover the situation regarding disorder, van der Waals contacts etc in the ‘Disorder’ field. At this point, it is left to the CSD user to decide for themselves.”
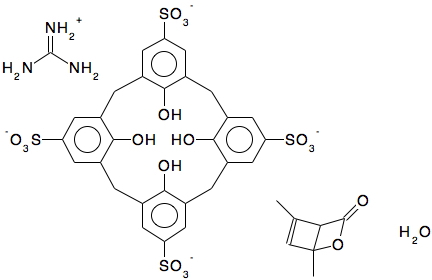
We have arrived at a point where the CSD user must indeed decide what the species described by this dataset actually is. Ideally, the best recourse would be to acquire the original data in full and repeat the crystallographic analysis. This is an aspect of the curation of crystallographic data that is not conducted as part of the current processes, which would require as a minimum a superset known as the hkl information to be present in the data. Again, to quote the CSD scientific editors:
- “With regard to your question: Is there any mechanism in the Conquest search to identify structures where the hkl information is present? I understand that it is not currently possible to do this in ConQuest. It is, however, possible … to access structure factor data (where available) using Access Structures.”
For MUWMEX, the hkl information is not present in the CSD dataset and in 2010 when the structure was published would have to be obtained directly from the authors. By 2016 however, its presence in deposited datasets was becoming far more common. It is worth pointing out that even the hkl information is not the complete data recorded for the experiment. That is represented by the original image files recording the X-ray diffractions. This latter is hardly ever available as FAIR data even nowadays.
I hope I have here illustrated at least some of the challenging aspects of curating scientific data and the issues that can arise when derived metadata (in this case the name and the atom connectivities of a molecule) reveal conflicts with the original interpretations. This for an area of chemistry where both the data deposition and its curation is a very mature subject, having operated for ~52 years now. It is still a process that requires the intervention of skilled curators of the data, but perhaps even more importantly it reveals the need to identify even more strictly what the provenance of the interpretations is. Should the CSD curation rest merely at the stage of teasing out and flagging inconsistencies and allowing the user to then take over to resolve the conflicts? Should it be more active, in re-analyzing data for each entry where conflicts have been detected? Perhaps the latter is not practical now, but it might be in the near future. What is certain is that with increasing availability of FAIR data these sorts of issues will increasingly come to the fore. And not just for the very well understood case of crystallographic data but for many other types of data.
References
- Y. Legrand, A. van der Lee, and M. Barboiu, "Single-Crystal X-ray Structure of 1,3-Dimethylcyclobutadiene by Confinement in a Crystalline Matrix", Science, vol. 329, pp. 299-302, 2010. https://doi.org/10.1126/science.1188002
- M.D. Wilkinson, M. Dumontier, I.J. Aalbersberg, G. Appleton, M. Axton, A. Baak, N. Blomberg, J. Boiten, L.B. da Silva Santos, P.E. Bourne, J. Bouwman, A.J. Brookes, T. Clark, M. Crosas, I. Dillo, O. Dumon, S. Edmunds, C.T. Evelo, R. Finkers, A. Gonzalez-Beltran, A.J. Gray, P. Groth, C. Goble, J.S. Grethe, J. Heringa, P.A. ’t Hoen, R. Hooft, T. Kuhn, R. Kok, J. Kok, S.J. Lusher, M.E. Martone, A. Mons, A.L. Packer, B. Persson, P. Rocca-Serra, M. Roos, R. van Schaik, S. Sansone, E. Schultes, T. Sengstag, T. Slater, G. Strawn, M.A. Swertz, M. Thompson, J. van der Lei, E. van Mulligen, J. Velterop, A. Waagmeester, P. Wittenburg, K. Wolstencroft, J. Zhao, and B. Mons, "The FAIR Guiding Principles for scientific data management and stewardship", Scientific Data, vol. 3, 2016. https://doi.org/10.1038/sdata.2016.18
Tags: assigned chemical name, author, chemical name, chemical name synonym, chemical names, chemical structures, editor, indicated chemical name synonym, Knowledge, radiation, Research, Scientific method, Technology/Internet, X-ray
I have been asked why I focus on the CSD (Cambridge structure database) which requires a commercial license to use, rather than the COD (Crystallography Open Database) which does not. Other than the size of the database (>800,000 vs 377077 sets of coordinates), the software tools for structure searching and analysis are much more complete and it has to be said easier to use for the former. What is more of an unknown is the level of curation applied to the structures in the database. So I thought it would be interesting to compare the curation for the structure discussed above with any entry in COD.
Unfortunately, the structure above has not been deposited in COD, and so no curation information about it is available. Neither, as it happens is the “silyl cation” which was described in this post.
I came across this quotation from the Retraction Watch site; “In view of … the essential absence of original data for the experiments reported in the article, and the widespread lack of clarity concerning how the experiments were conducted, it is the opinion of the Expert Group that the article in Science should be recalled.” which is typical of the importance of original data now being attributed.
Here is another example illustrating the importance of having access to original research data. The Comment on “Crystallographic Snapshot of an Arrested Intermediate in the Biomimetic Activation of CO2” (DOI: 10.1002/anie.201411654) has the following quote, referring to crystallographic data:
“However, since we have no access to the original crystallographic data, ….”
They instead used computational geometry optimisations on the claimed structures reported at DOI: 10.1002/anie.201407165 (a corrigendum is at 10.1002/anie.201504197) and conclude with the statement:
In light of the crystallographic and computational data reported herein, we believe that the crystal structure of [nBu4N][O2C⋅⋅⋅OH] reported by Ackermann et al. is in fact that of [nBu4N][O2CCH3].

BUT. Given that they base their arguments, at least in part, on computed coordinates, the supporting information provided with the article 10.1002/anie.201411654 has remarkably non-FAIR computational data of its own! Thus figures S15-S18 in this SI does not include FULL coordinates for the systems discussed, nor does it include any other computational outputs which would allow one to check on the integrity of the calculations.
I am here following up on my previous comment about the absence of crystallographic data which would enable a closer look at a crystal structure purportedly reporting the capture of the –O2C-OH anion.
How about mining the CSD for examples of this species? The search query to do this is available at DOI: 10.14469/hpc/2537 and this yields the result:
Note that that the original report of this species (DOI: 10.1002/anie.201407165) indicated a HO…C bond length of ~1.563Å. The 33 hits (no errors, no disorder, R < 0.05) indicate there are no examples even close to this value, very strongly suggesting it is in error. This error was indeed recognised in the form of a corrigendum shortly after the original publication; 10.1002/anie.201504197.
At this point one has to ask why the referees of the original article did not run the same check as above, since it only takes ~5 minutes to perform!
This is a recently published article in the journal of chemical education, DOI: 10.1021/acs.jchemed.5b00629 which introduces the cyclobutadiene example and several other crystal structures for discussion in undergraduate classes.