The idea of so-called FAIR (Findable, Accessible, Interoperable and Reusable) data is that each object has an associated metadata record which serves to enable the four aspects of FAIR. Each such record is itself identified by a persistent identifier known as a DOI. The trick in producing useful FAIR data is defining what might be termed the “granularity” of data objects that generate the most readily findable and which most usefully enable the other three attributes of FAIR.
To set the scene for how to do this optimally, I first set out two extreme examples of FAIR objects relating to chemical spectroscopy such as NMR. These will be directly associated with a journal article describing for arguments sake say 50 compounds new to science, with the existence of these data objects identified via a data availability statement appended to the article. Each compound might be characterised by say spectroscopic and crystallographic information and perhaps some computational analysis. For the spectroscopic analysis, perhaps 5 types of NMR experiments might be included, giving a total of around 10 separate types of datasets for each compound, or in round numbers lets say 500 data sets for the 50 compounds reported in such an article.†
- Method A: The data associated with an articles takes the form of a ZIP (or other type of compressed) archive containing all 500 of the intended FAIR data sets. The resulting ZIP file is then described with a single metadata record and assigned a single DOI using e.g. the tools of a data repository. That one metadata record has the (mammoth) task of describing all of these datasets, across perhaps ten different kinds of experiment. This type of monolithic object is in fact not unusual, for several reasons. Some repositories impose a significant charge for each deposition, and so the temptation to reduce costs would be to adopt this expedient.
- Method B: The other extreme is to literally deposit all 500 data sets separately and assign 500 DOIs, each with a separate metadata record. The issue now is less how well the metadata record can describe each dataset, but more of to establish the relationships between these 501 objects (the journal article and each dataset). Such relationships could include:
- that between the compound molecular structure and the dataset
- that between say the dataset and the type of spectroscopic experiment (e.g. IR, MS, NMR, XRD, Comp)
- that between different eg NMR experiments for the same compound (the nucleus, the pulse sequence, the solvent, etc).
- These could in total represent a great many individual relationships between both the 500 data sets and the article itself (formally around 5012/2!)
Before setting our solution, I show below how a typical repository such as Zenodo handles the relationships between data objects noted above.
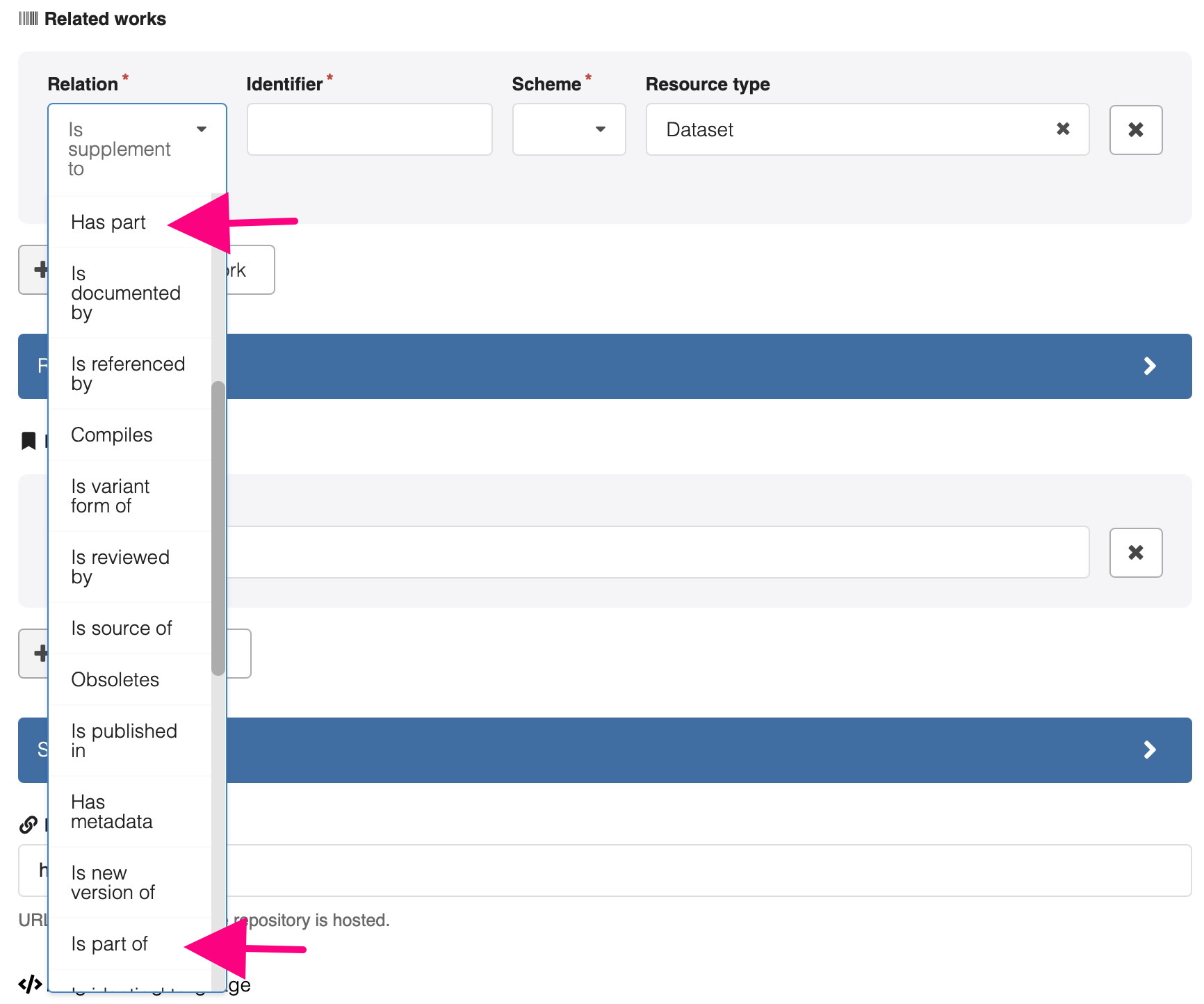
The relation type is selected from a controlled list of about 30, and is entered for each individual metadata record associated with a DOI. So clearly, relationships in the second category would have to be individually entered, hardly feasible for 5012/2 entries. And in the first category, only one relationship between the single large archive of data and the journal DOI can be added. One of the more important relationships in this context are the “Has part” or “Is part of” ones (diagram above).
The use of this now constitutes Method C.
- One starts by creating what could be called a top or level 1 entry, which will contain important core metadata information such as the contributing authors, the institute where the data was obtained, the title and overall description of the datasets to come, a license, a date, a declaration of the published article associated with the data and finally the DOI of this metadata record. This top-level entry would also list all the compounds on level 2 for which data is available and each being referenced by a “Has part” declaration via a DOI for each compound.
- Each compound on level 2 would in turn point back to level 1 by an “Is part of” metadata declaration. Each compound on level 2 would also list the spectroscopic experiments available that compound, for example the NMR method as part of level 3. It would have an “Is part of” declaration pointing back to the compound level 2 entry.
- The list of the different NMR experiments on level 3 also have “Has part” declarations pointing to the list of NMR experiments on level 4.
- Each NMR experiment conducted on level 4 would contain an “Is part of” declaration back to level 3 and a list of “Has part” entries which describe the individual data files available for that experiment in the metadata record for level 4.
If you wish, you can inspect all “Has part”/”Is part of” declarations in the metadata records for these various levels by invoking e.g. https://data.datacite.org/application/vnd.datacite.datacite+xml/10.14469/hpc/11446 (replacing e.g. 11446 by any of the DOI suffixes shown in red in the diagram below). They are all associated with this published article.[1]
What does this use of relational parts declarations achieve? Well, compared to method A, where everything had to be achieved within a single metadata record (and in practice never is) or method B, where a very large number of relationships would have to be declared (and again never are), Method C achieves a good balance between the two.‡ By collecting the metadata information into groups, one can achieve a more readily navigable structure for the information and also allow sub-groups to effectively inherit properties from the higher group.
I end by noting that far too few FAIR data collections associated with published journal articles adopt such procedures, in large part because of very little current exploitation of relationships between the data such as the one used above (“Has part”/”Is part of”). The repository itself has to be carefully designed to do this as automatically as possible and not require the human depositor to invoke each instance by hand (as shown for e.g. Zenodo above). An example of just such a repository is described here.[2]
†The data sets themselves might be made available in more than one form (for NMR, a Bruker ZIP archive, an Mnova file, a JCAMP-DX format or just a PDF spectrum), thus increasing the number even further.
‡It reminds me of when I used to teach molecular orbital theory using the Hückel method, which requires a secular matrix to be diagonalised. For e.g. naphthalene, this operation would have to be conducted on a 10*10 matrix, something almost impossible by hand. However, one could use group theory to block diagonalise this matrix into much smaller matrices with the off-diagonal elements between them set to zero, thus considerably reducing the task at hand.
References
- T. Mies, A.J.P. White, H.S. Rzepa, L. Barluzzi, M. Devgan, R.A. Layfield, and A.G.M. Barrett, "Syntheses and Characterization of Main Group, Transition Metal, Lanthanide, and Actinide Complexes of Bidentate Acylpyrazolone Ligands", Inorganic Chemistry, vol. 62, pp. 13253-13276, 2023. https://doi.org/10.1021/acs.inorgchem.3c01506
- M.J. Harvey, A. McLean, and H.S. Rzepa, "A metadata-driven approach to data repository design", Journal of Cheminformatics, vol. 9, 2017. https://doi.org/10.1186/s13321-017-0190-6